The LIFS consortium provides regular in-person or virtual training events at the Lipidomics Forum conference and at other conferences. We also offer online, self-paced training material at our Moodle site.
- Hits: 563

The LIFS consortium provides regular in-person or virtual training events at the Lipidomics Forum conference and at other conferences. We also offer online, self-paced training material at our Moodle site.

Date & Time:
March 24-28, 2025
Registration Deadline:
February 10, 2025
Topic: Bioinformatics for Lipidomics, with a focus on mass spectrometry data analysis and workflows
Contents: The de.NBI Spring School 2025 will provide in-depth knowledge and practical experience in lipidomics bioinformatics. Participants will explore state-of-the-art bioinformatics tools essential for lipidomics research, comprehensive lipidomics workflows, and the challenges and limitations of lipidomics from multiple perspectives.
Prerequisites:
Basic knowledge of lipidomics, analytical workflows in lipidomics, and basic familiarity with web-based and desktop applications. Please note that you may need to install software on your computer to fully participate in all exercises requiring proper rights. The workshop will be a mix of small lecture segments and hands-on exercises. The trainers will be available for questions and assistance during the workshops.
Learning goals: Gain understanding of bioinformatics for lipidomics, possibilities,
challenges, and limitations
Keywords:
Lipidomics, Pre-Analytics, Research Data Management, Mass Spectrometry, Data Analysis, Visualization
Tools:
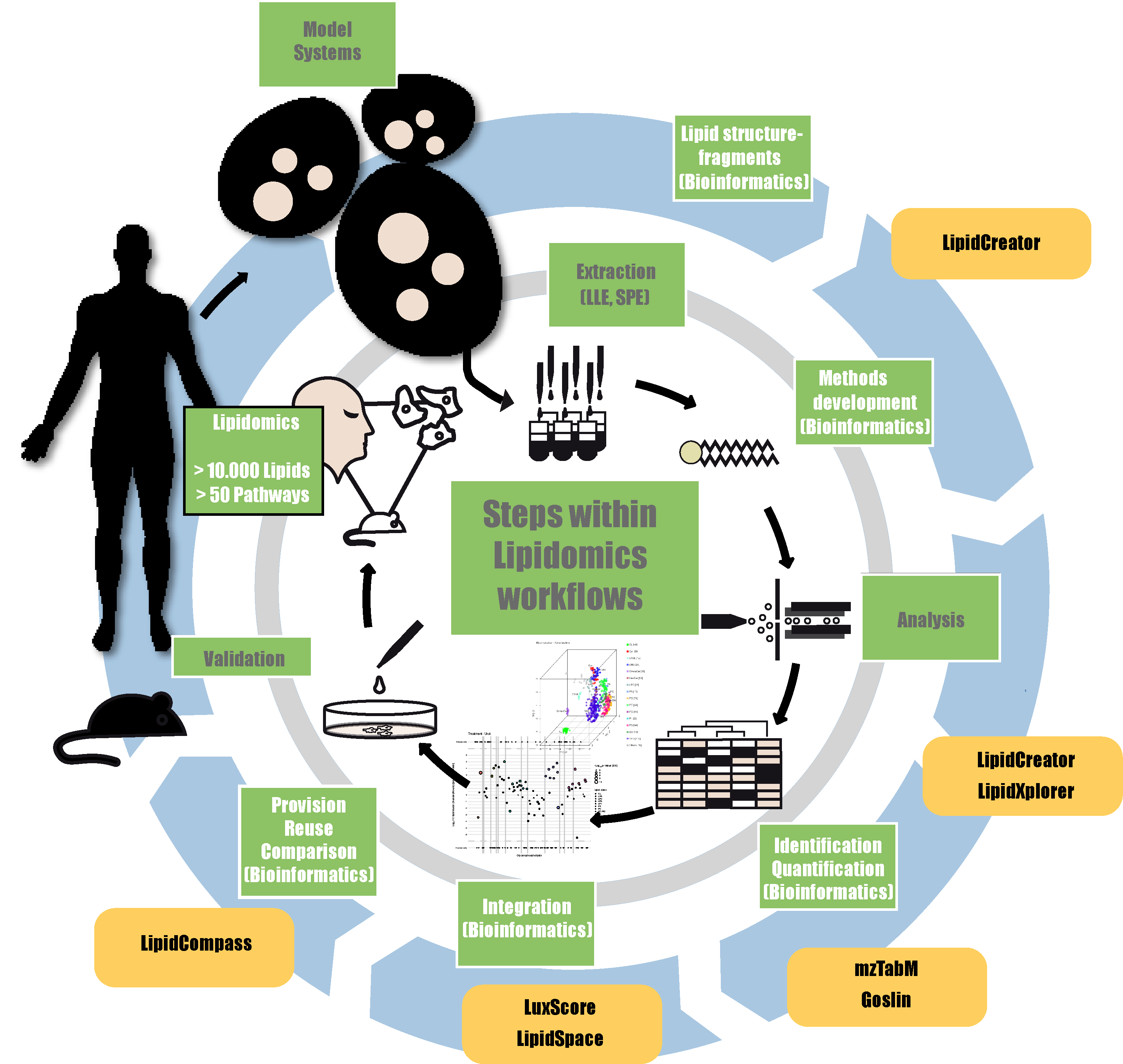
LipidCreator & Skyline, LipidXplorer & lxPostman, mzmine, LipidSpace, Lipidomics Checklist, Goslin, LipidCompass, MetaboLights
Organizers:
Trainers:
Keynote Speakers:
Location:
On-Site, Center for Biotechnology, Bielefeld University, Bielefeld, Germany
Accommodation: For accepted participants, accommodation costs will be covered by the
organizers.
More information and application:
https://www.denbi.de/training-courses-2025/1847-de-nbi-spring-school-2025-
bioinformatics-for-lipidomics
Contact:
Please use our contact form if you have any questions!
This workshop is kindly supported by the German Network for Bioinformatics Infrastructure (de.NBI) and Elixir Germany:
Educators:
Robert Ahrends (Uni Vienna, LIFS), Dominik Kopczynski (Uni Vienna, LIFS), Nils Hoffmann (FZ Jülich, LIFS), Dominik Schwudke (FZ Borstel, LIFS)
Location:
On-Site training as part of 9th Lipidomics Forum 2024 in Borstel.
Date & Time:
Sunday, September 1st 2024
10:00 - 16:30
Contents:
In the first part of the course, we will work through an example of targeted LC-MS lipidomics with LipidCreator and Skyline. In the second part of the course, we will provide insights into using LipidXplorer to identify and quantify lipids from shotgun lipidomics. The course will consist of a short theory and background overview of the employed programs complemented by applying the tools to provided data sets. The third part of this course will work through a typical use-case of downstream data processing of shotgun lipidomics data following MS acquisition with LipidXplorer. We will inspect, check, and normalize the data and calculate absolute quantities using internal class-specific standards with lxPostman. We will then perform a qualitative and quantitative comparison of the lipidomes using LipidSpace.
Learning goals:
Participants will be able to understand and explain the targeted shotgun MS and LC-MS workflows for lipidomics. They will learn the fundamentals of the software tools used and how to choose parameters for them. They will learn to understand and interpret the results of each pipeline step.
Prerequisites:
Basic knowledge of lipidomics, analytical workflows in lipidomics, and basic familiarity with web-based and desktop applications. Please note that you may need to install software on your computer to fully participate in all exercises requiring proper rights. The workshop will be a mix of small lecture segments and hands-on exercises. The trainers will be available for questions and assistance during the workshop.
Keywords:
Lipidomics, Shotgun, Targeted Analysis, LC-MSn, Reporting
Tools:
LipidXplorer, LipidCreator, Skyline, Goslin, lxPostman, LipidSpace, Reporting Checklist
Registration:
Please register for the course by ticking the LIFS Workshop option on the Lipidomics Forum registration page. (Fee: 20 €)
We will contact you prior to the course with instructions to access the Moodle course material.
Contact:
Please use our contact form if you have any questions!
This workshop is kindly supported by the German Network for Bioinformatics Infrastructure (de.NBI) and Elixir Germany:
Page 1 of 3