Welcome to the web portal for Lipidomics Informatics for Life Sciences (LIFS)

Even with lipid content of over 10% in the human body, lipids were not the central scientific focus during the last decade. However, more and more evidence proves that non-genetically determined molecules such as metabolites and lipids play a critical role in cellular regulation. Today, it is evident that lipids are not only crucial for energy homeostasis and as an environmental-cellular barrier but also that they represent a central part of our signal transduction machinery.
Disruptions of the sensitive lipid metabolism are highly correlated with different diseases, including thrombocytopenia, metabolic syndrome, diabetes, obesity, and hyperlipidemia. This is especially true for the latter, which also reach pandemic levels. Therefore, lipid metabolism is becoming an emerging scientific field and is a central part of pharmacological research today (statins, cyclooxygenase inhibitors, endurobol in doping). Thus, the mission of the LIFS is to provide the bioinformatic framework to understand lipids in context and create an integrative systems biology view for lipid research.
de.NBI and LIFS
Lipidomics encompasses analytical approaches that aim to identify and quantify the 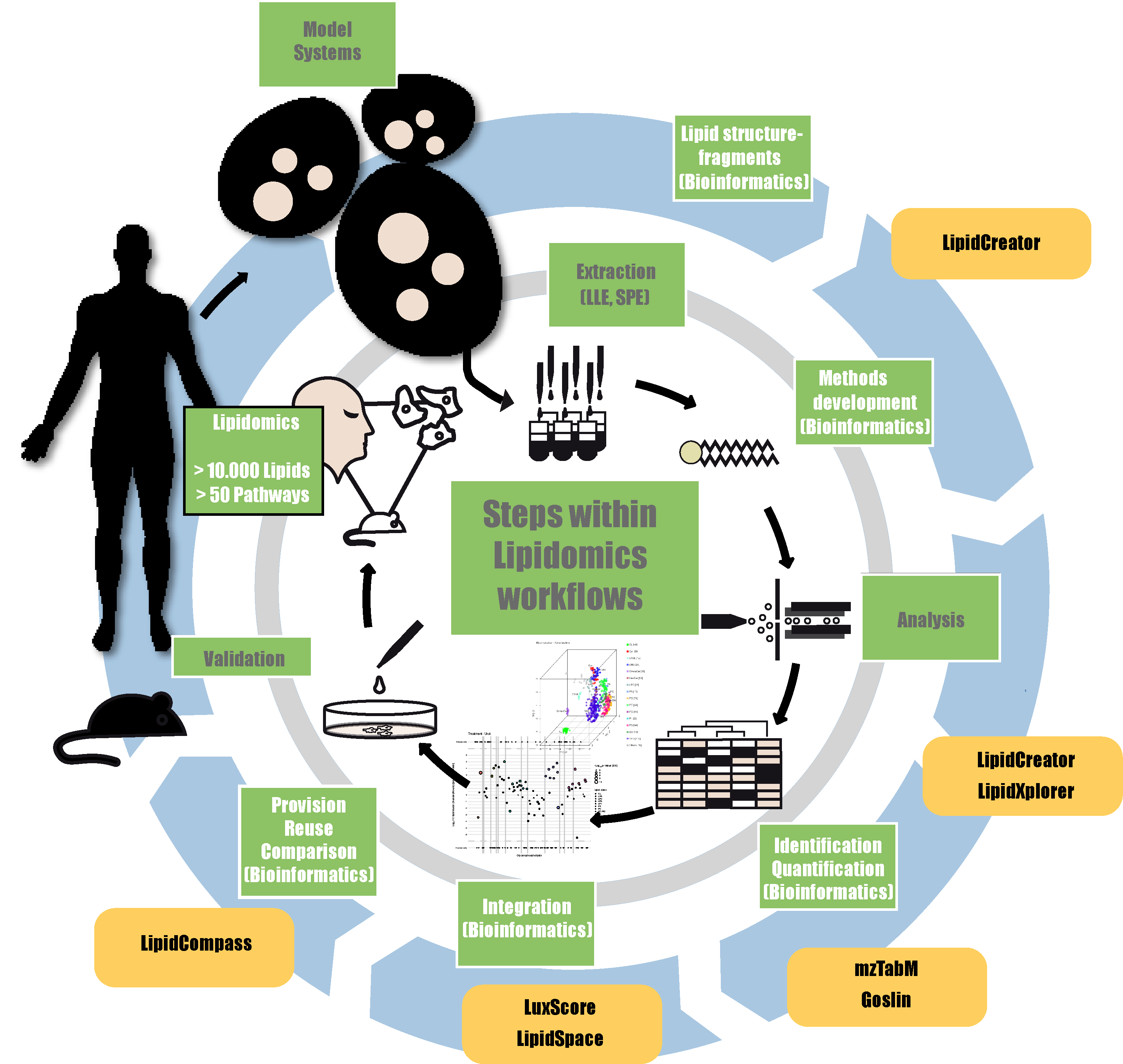 complete set of lipids, defined as the lipidome in a given cell, tissue, or organism and their interactions with other molecules. Most lipidomics workflows are based on mass spectrometry and have been proven as a powerful tool in system biology in concert with other Omics disciplines. Unfortunately, bioinformatics infrastructures for this relatively young discipline are limited only to some specialists. Services, search engines, quantification algorithms, visualization tools, and databases developed by the 'Lipidomics Informatics for Life-Science' (LIFS) partners will be established and hosted via the LIFS portal to provide broad access to these specialized bioinformatics pipelines.
complete set of lipids, defined as the lipidome in a given cell, tissue, or organism and their interactions with other molecules. Most lipidomics workflows are based on mass spectrometry and have been proven as a powerful tool in system biology in concert with other Omics disciplines. Unfortunately, bioinformatics infrastructures for this relatively young discipline are limited only to some specialists. Services, search engines, quantification algorithms, visualization tools, and databases developed by the 'Lipidomics Informatics for Life-Science' (LIFS) partners will be established and hosted via the LIFS portal to provide broad access to these specialized bioinformatics pipelines.
The capacity building suggested by LIFS will dissect the molecular foundation of many different lipid-related dysfunctions. LIFS is a member of the 'German Network for Bioinformatics' (de.NBI), and the LSI (Lipidomics Standard Initiative) and will provide access to the described software as well as to tutorials and consulting services for lipidomics research and applications via the LIFS web-portal.
In particular, we develop and maintain the following software tools:
- LipidCreator - lipidomics assay design and library generation for targeted lipidomics, integrated with Skyline
- LipidXplorer - shotgun lipidomics, identification, and quantification
- LUX Score - qualitative lipidome comparison based on a measure of structural homology between lipidomes similar to genetic analyses
- Clover - quantitative, multi-factorial visualization of lipidomics data, statistical comparison
- LipidCompass - quantitative lipidomics experiment database to compare lipid levels across studies, samples, tissues, and species
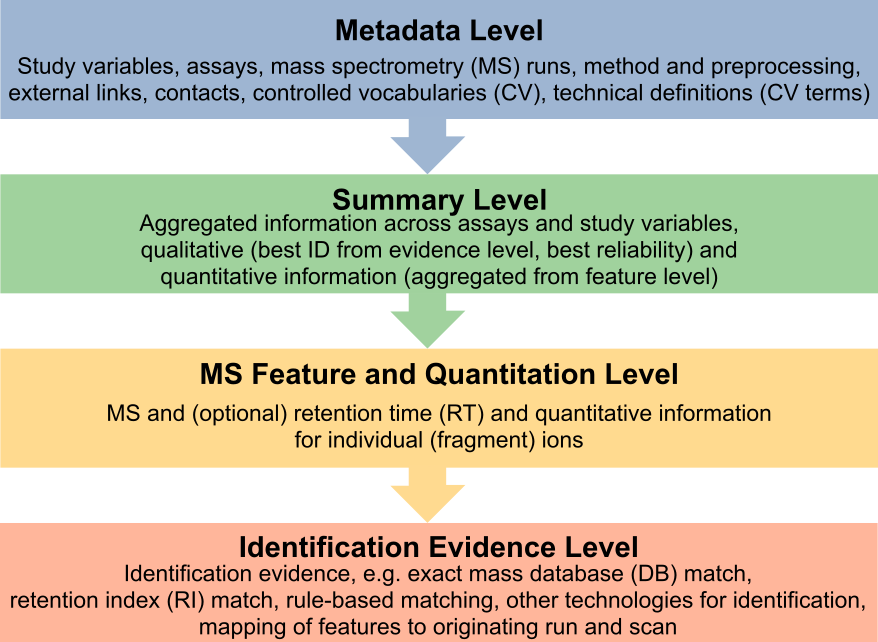
Standardization in Lipidomics
LIFS collaborates with the Lipidomics Standards Initiative on standardization efforts concerning lipid identification, quantification, naming and reporting.
The mzTab-M: a data standard for sharing quantitative results in mass spectrometry metabolomics is a candidate to become the format of choice to report results from quantitative lipidomics experiments.
More information on mzTab-M for lipidomics is available at our GitHub project.
The organizers:
Dominik Schwudke, Andrej Shevchenko, Robert Ahrends, Nils Hoffmann
Current funding
Continued operation of LIFS is supported by:University of Vienna, since Jan. 1st, 2022.
Forschungszentrum Jülich, since Jan. 1st, 2022.
This work is funded by the Federal Ministry of Education and Research in the frame of de.NBI & ELIXIR-DE (Forschungszentrum Jülich and W-de.NBI-006) since Jan. 1st, 2023.
Former funding
History
The Leibniz Institut für Analytische Wissenschaften-ISAS e.V.- was a partner of the LIFS project until Dec. 31st 2021.
Ruhr-Uni Bochum was the de.NBI service center and partner of the LIFS project until Dec. 31st 2021.
Please also see our terms of service.






