User Survey:
In order to improve our services, please provide your feedback in our Lipidome Projector.
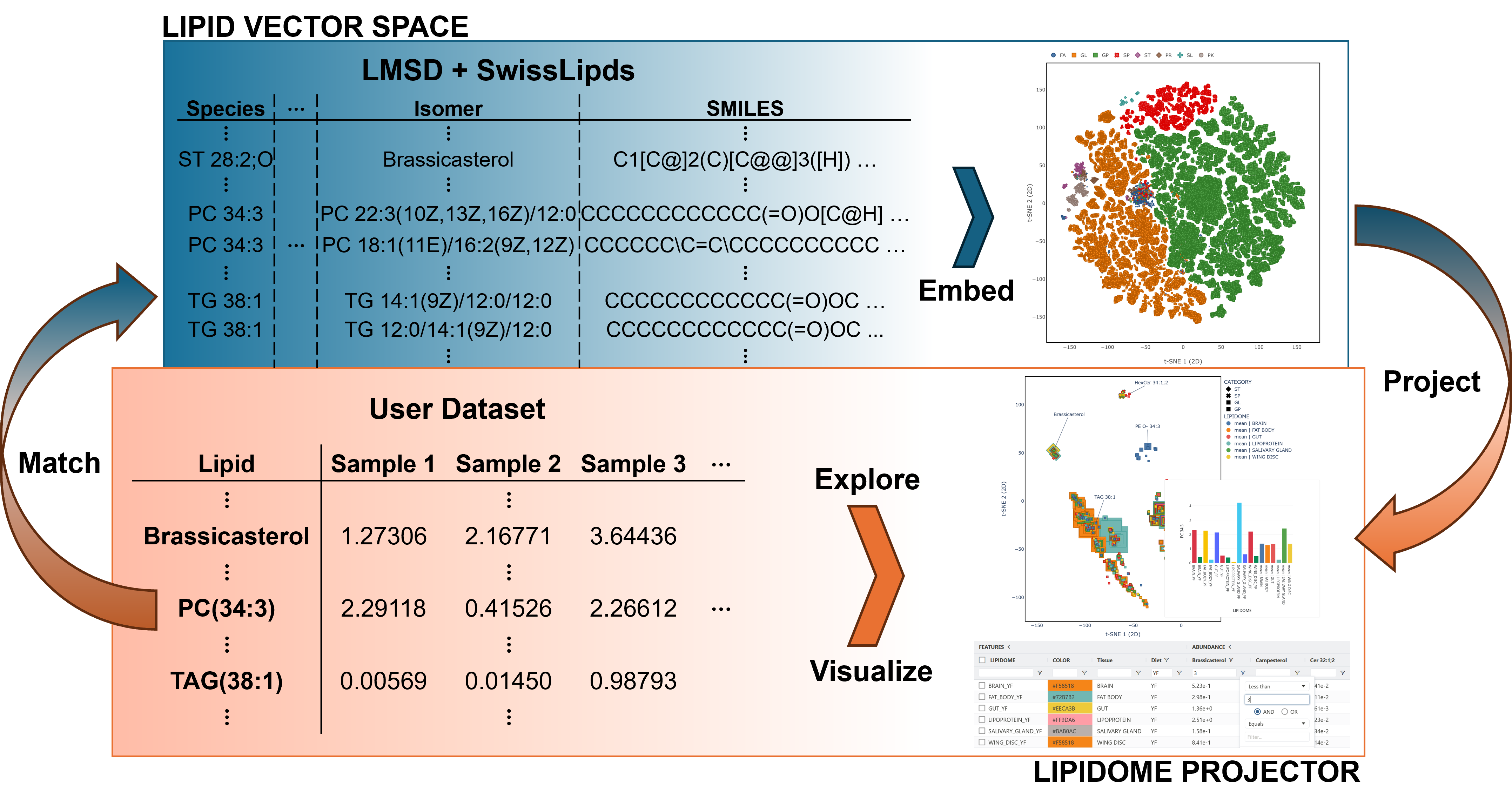
Lipidome Projector
Lipidome Projector is a web-based tool for the interactive visualization, exploration and analysis of lipidomics datasets.
A neural network was used to compute a vector space for a large set of lipid structures from the Lipid Maps Structure Database [1] and SwissLipids [2].
User lipidomes are matched by notation, projected onto this space, and displayed as 2D / 3D scatter plots. This allows a quick overview over the distributions of abundances across lipidome samples and lipid classes.
Fully interactive plots, data grids and data processing operations (sorting, filtering, averaging of samples and fold change computation) allow an interactive exploration of datasets to find and display structural patterns in lipid abundances.
Availability
Lipidome Projector is in active development. It is an open-source Python package available under the MIT license.
A preliminary test-server is available, where users may upload and analyse their own lipidomics data, or familiarize themselves with the software by exploring one of the development datasets [3,4,5].
The methods are described in a preprint [6] and the corresponding journal article is in revision.
References
[1] Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35: D527--D532. doi:10.1093/nar/gkl838
[2] Aimo L, Liechti R, Hyka-Nouspikel N, Niknejad A, Gleizes A, Götz L, et al. The SwissLipids knowledgebase for lipid biology. Bioinformatics. 2015;31: 2860--2866. doi:10.1093/bioinformatics/btv285
[3] Carvalho M, Sampaio JL, Palm W, Brankatschk M, Eaton S, Shevchenko A. Effects of diet and development on the Drosophila lipidome. Mol Syst Biol. 2012;8. doi:10.1038/msb.2012.29
[4] Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106. doi:10.1073/pnas.0811700106
[5] Lunding LP, Krause D, Stichtenoth G, Stamme C, Lauterbach N, Hegermann J, et al. LAMP3 deficiency affects surfactant homeostasis in mice. PLoS Genet. 2021;17. doi:10.1371/journal.pgen.1009619
[6] Olzhabaev T, Müller L, Krause D, Schwudke D, Torda AE. Lipidome visualisation, comparison, and analysis in a vector space. bioRxiv 2024, doi:10.1101/2024.08.02.606134
Contact and Help Desk
For any further information, please contact us.
