Organizers:
de.NBI Service Center BioInfra.Prot, Lipidomics Informatics for Life Science
Nils Hoffmann, Helena Schnitzer, Daniel Wibberg (FZ Jülich)
Dominik Schwudke (Research Center Borstel)
Dominik Kopczynski, Robert Ahrends (University of Vienna, Austria)
Educators:
Daniel Wibberg, Sebastian Beier, Sara Beier, Nils Hoffmann (all FZ Jülich)
Dominik Kopczynski (University of Vienna)
Lisa Hahnefeld (Goethe-Universität Frankfurt und Fraunhofer ITMP)
Dominik Schwudke, Ballal Hossen (FZ Borstel)
Ansgar Korf (mzmine)
Noemi Tejera Hernandez (EMBL-EBI, MetaboLights)
Lisa Hahnefeld (Goethe-Universität Frankfurt und Fraunhofer ITMP)
Cristina Coman & Nina Troppmair (Skyline & LipidCreator Tutorial Material, University of Vienna)
Keynotes:
Sebastian Beier & Sara Beier
Andrej Shevchenko / Vannuruswamy Garikapati
Robert Ahrends
Hanna Bednarz
Date & Time:
Monday, March 24th - Friday, March 28th 2025
Contents:
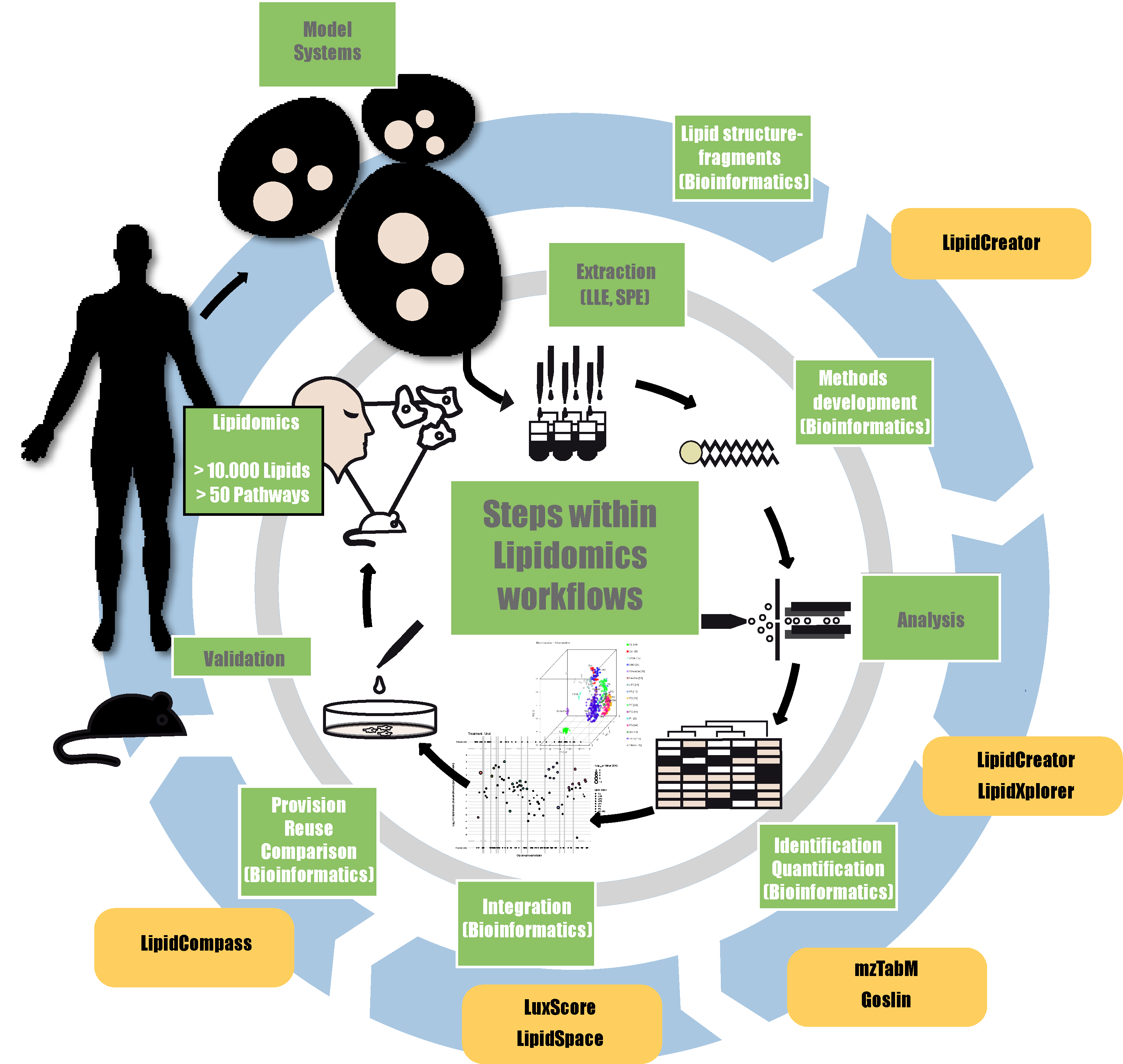
The LIFS de.NBI partner project has pioneered innovative software solutions in the field of mass spectrometry for lipidomics data analysis. Building on this expertise, the de.NBI Spring School 2025 will concentrate specifically on Lipidomics bioinformatics, providing participants with in-depth knowledge and practical experience in this dynamic field.
Throughout the training, attendees will explore state-of-the-art bioinformatics tools essential for Lipidomics research. They will be introduced to comprehensive lipidomics workflows, offering a clear understanding of the capabilities, challenges and limitations inherent in Lipidomics from multiple perspectives. This knowledge is crucial for effectively analyzing lipid data and integrating it with other biological data types.
The de.NBI Spring School 2025 will feature a mix of engaging lectures and interactive hands-on sessions led by experts from the de.NBI and ELIXIR networks and beyond. The program is designed for PhD students, postdoctoral fellows, and other scientists who have a foundation in bioinformatics and molecular biology, and who are eager to expand their expertise in Lipidomics bioinformatics.
Participants will leave equipped with the skills and insights necessary to advance their research and contribute to the growing field of Lipidomics, helping to unlock biological insights that can drive innovative solutions in health and disease management and other application areas.
Learning goals:
Participants will be instructed in Lipidomics workflows and gain understanding about the possibilities, challenges and limitations of Lipidomics.
Prerequisites:
The Spring School is targeted towards early career scientists like PhD students and Postdocs, but also experienced scientists, who want to learn more about Bioinformatics for Lipidomics. Participants should have basic knowledge of lipidomics, analytical workflows in lipidomics, and basic familiarity with web-based and desktop applications. Please note that you may need to install software on your computer to fully participate in all exercises requiring proper rights. The workshop will be a mix of small lecture segments and hands-on exercises. The trainers will be available for questions and assistance during their workshop sessions.
Keywords:
Lipidomics, Pre-Analytics, Research Data Management, Mass Spectrometry, Data Analysis, Visualization
Tools:
Alister, LipidXplorer, mzmine, LipidCreator, Skyline, Goslin, lxPostman, LipidSpace, Reporting Checklist, LipidCompass, MetaboLights
- Teacher: Sebastian Beier
- Teacher: Lisa Hahnefeld
- Teacher: Nils Hoffmann
- Teacher: Dr. Dominik Kopczynski
- Teacher: Dominik Schwudke
- Teacher: Noemi Tejera
- Teacher: Daniel Wibberg