LIFS Tools & Resources
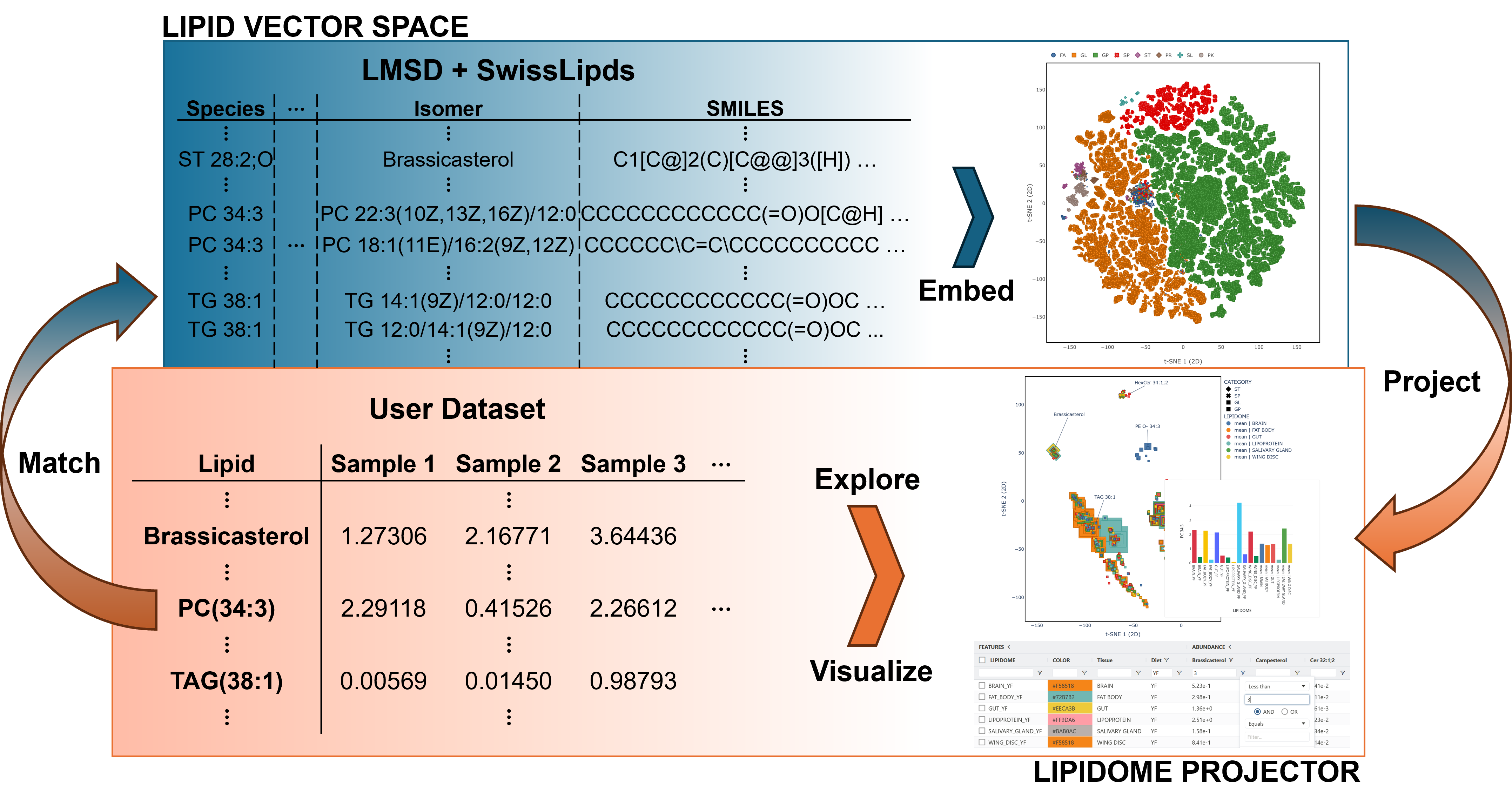
Lipidome Projector
Lipidome Projector is a web-based tool for the interactive visualization, exploration and analysis of lipidomics datasets. It uses a neural network to compute a vector space for a large set of lipid structures from the LIPID MAPS and SwissLipids databases and provides comprehensive plots and tools for interactive data exploration.
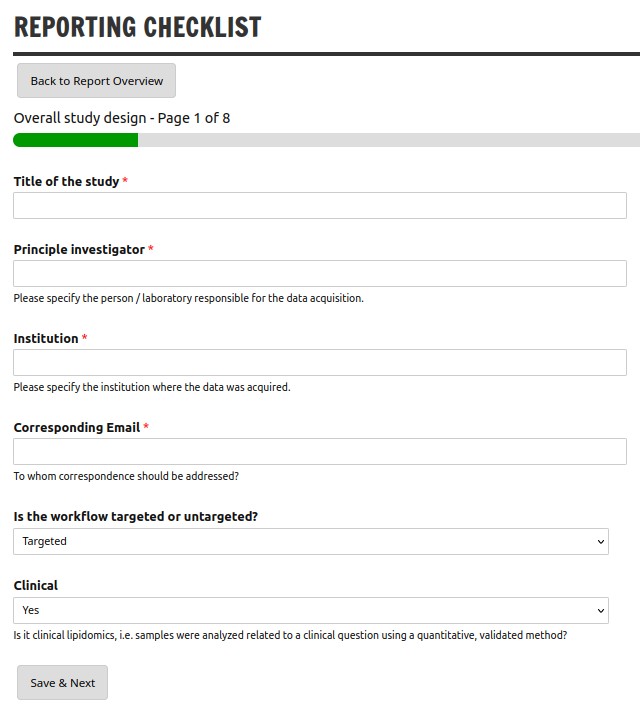
Reporting Checklist
The Lipidomics Minimal Reporting Checklist aims to standardize lipidomics research by providing guidelines for transparent, reliable, and reproducible data reporting across the field. The web application provides a convenient, step-wise wizard, export to PDF and optional creation of a DOI via Zenodo.
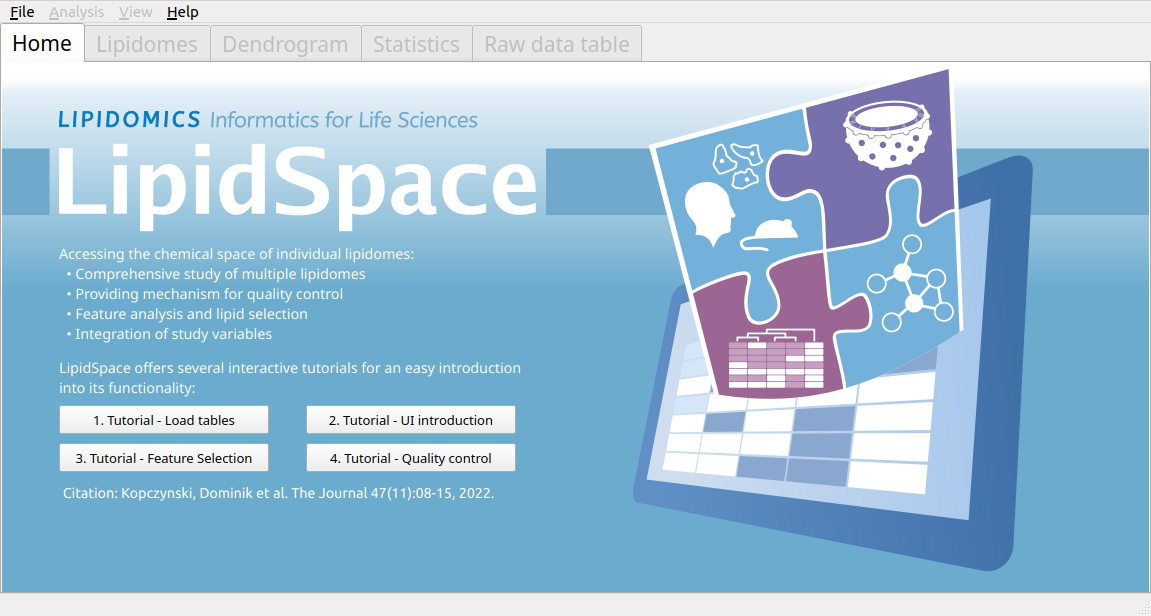
LipidSpace
LipidSpace is a stand-alone tool to analyze and compare lipidomes by assessing their structural differences. A graph-based comparison of lipid structures that allows to calculate distances between lipids and to determine similarities across lipidomes. It allows for a rapid (re)analysis of experiments, identifies lipids responsible for shaping the respective lipidome, and provides methods for quality control.
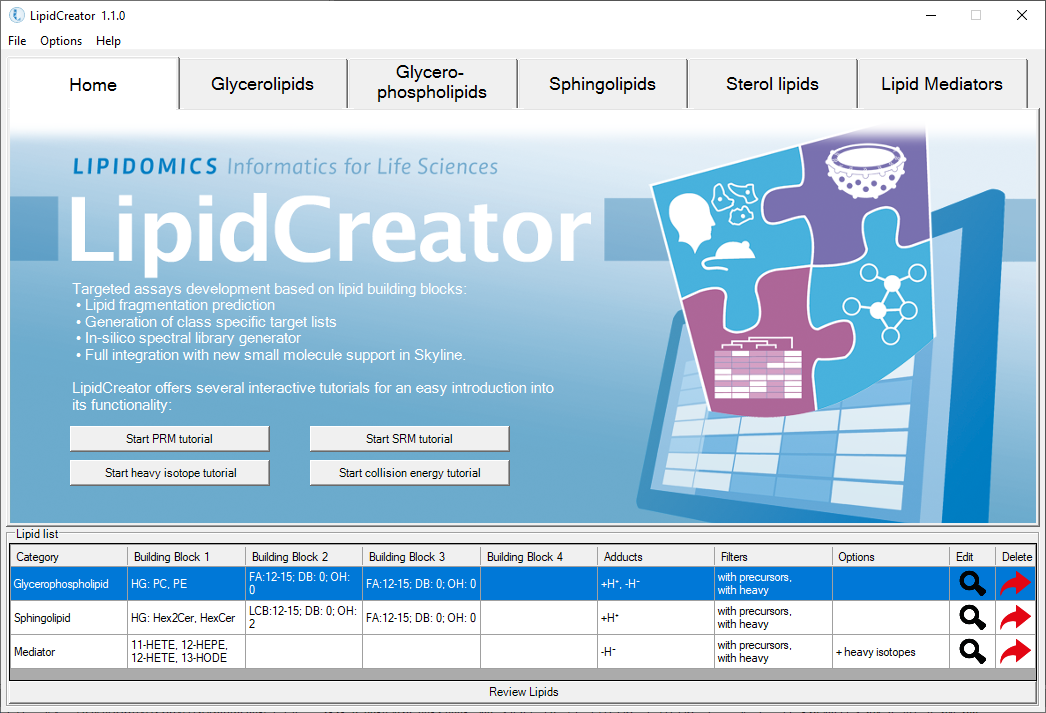
LipidCreator
LipidCreator is an advanced, user-friendly lipidomics platform designed to streamline the creation of targeted mass spectrometry assays. It supports a wide range of lipid categories and classes, integrates seamlessly with tools like Skyline, and offers powerful features such as spectral library generation and collision energy optimization, making it a versatile solution for comprehensive lipidomics research.
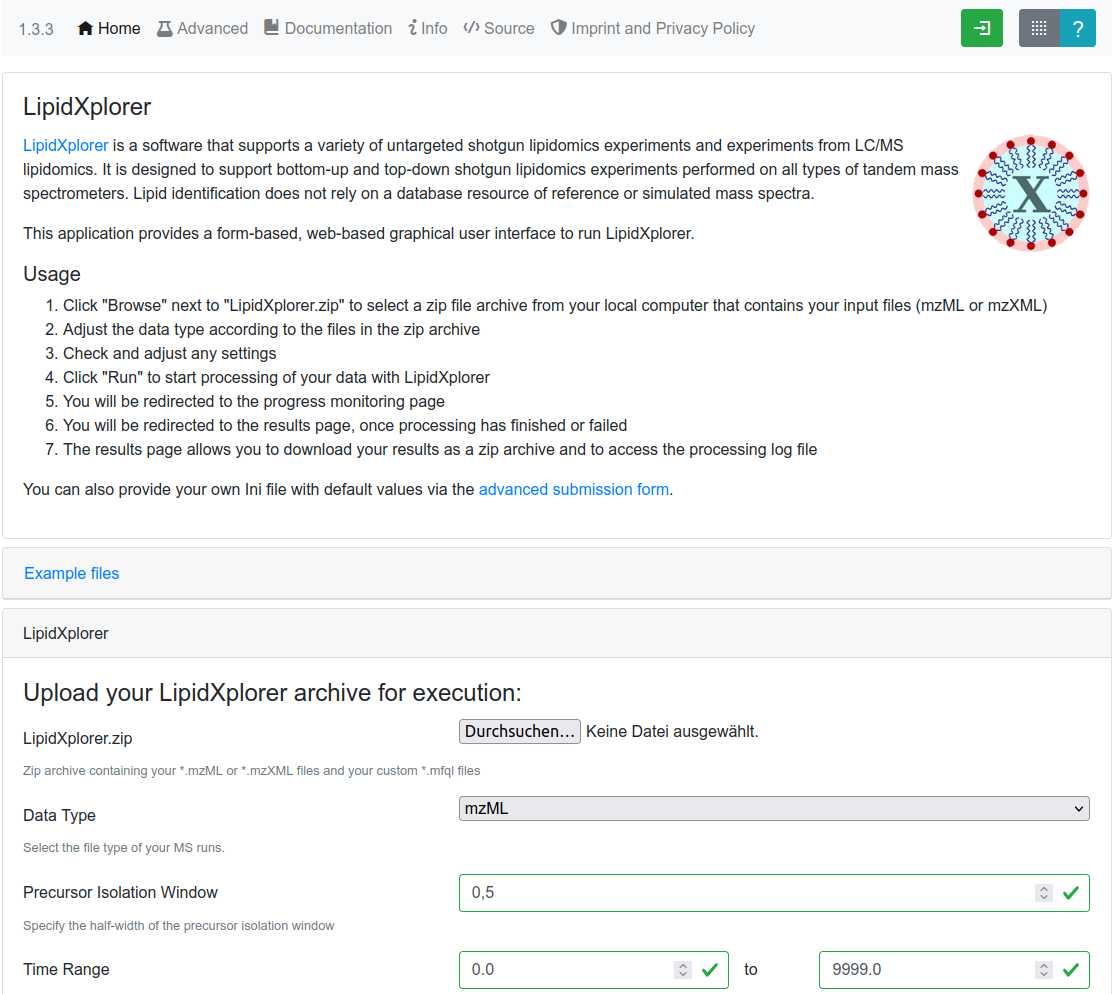
LipidXplorer
LipidXplorer is a software that supports a variety of untargeted shotgun lipidomics experiments and experiments from LC/MS lipidomics. It is designed to support bottom-up and top-down shotgun lipidomics experiments performed on all types of tandem mass spectrometers. Lipid identification does not rely on a database resource of reference or simulated mass spectra but uses the SQL-inspirece 'molecular fragmentation query language' MFQL.
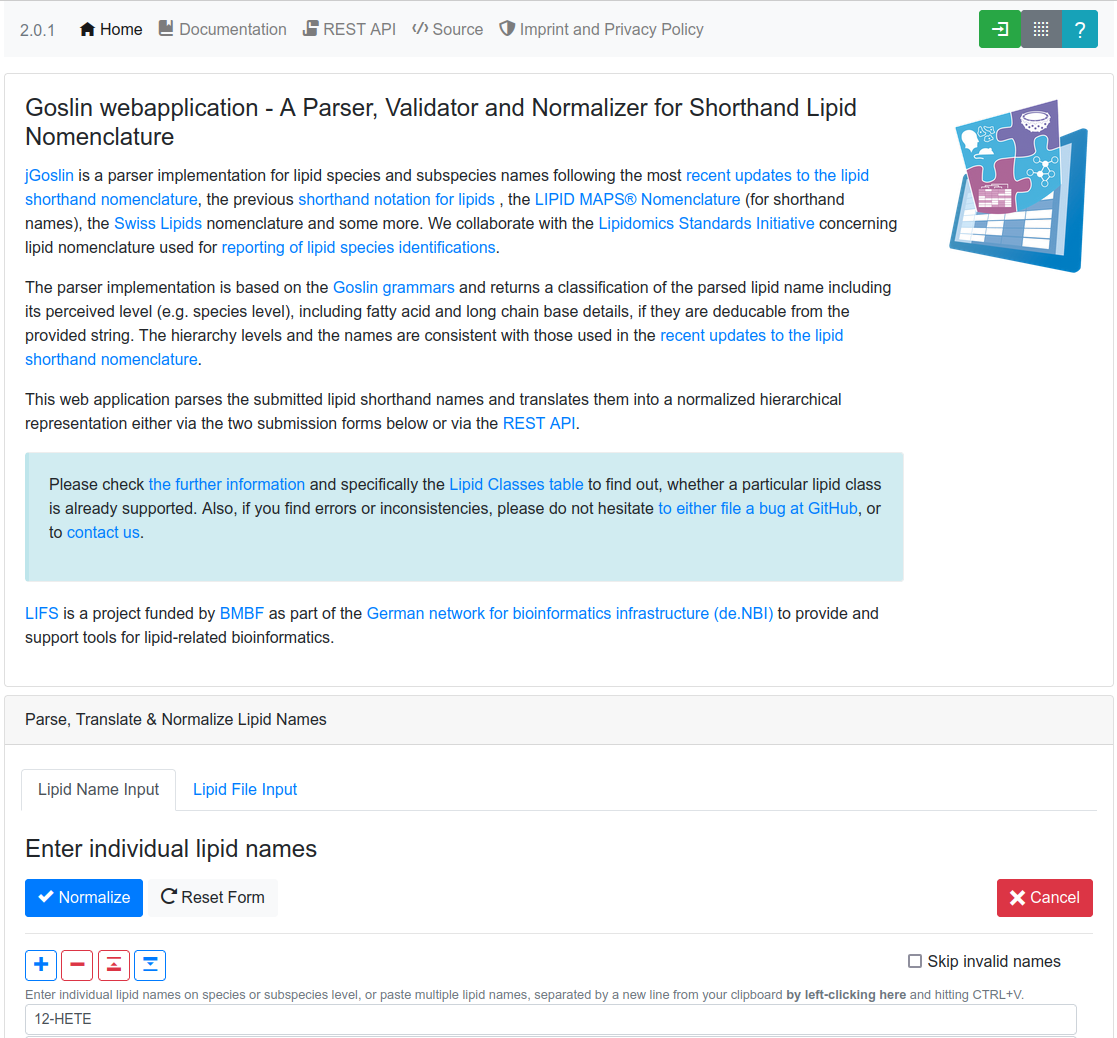
Goslin
Goslin can read different lipid nomenclatures. It automatically converts lipid names to the lipid shorthand nomenclature used by LIPID MAPS. The Goslin WebApp also provides mappings to LIPID MAPS and SwissLipids entries using the normalized shorthand name of lipids.
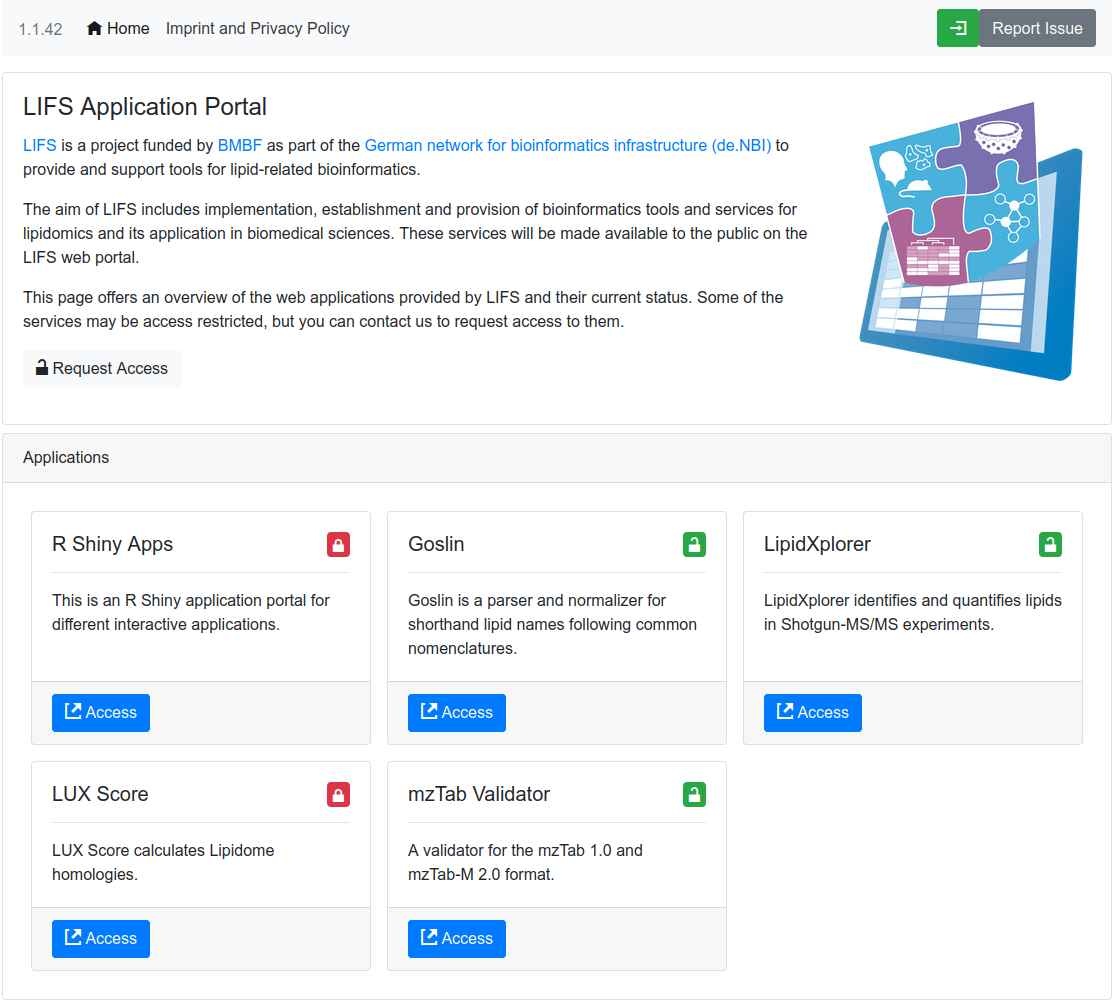
LIFS Web Applications
The LIFS Web Application portal provides an entry point to multiple LIFS lipidomics tools.
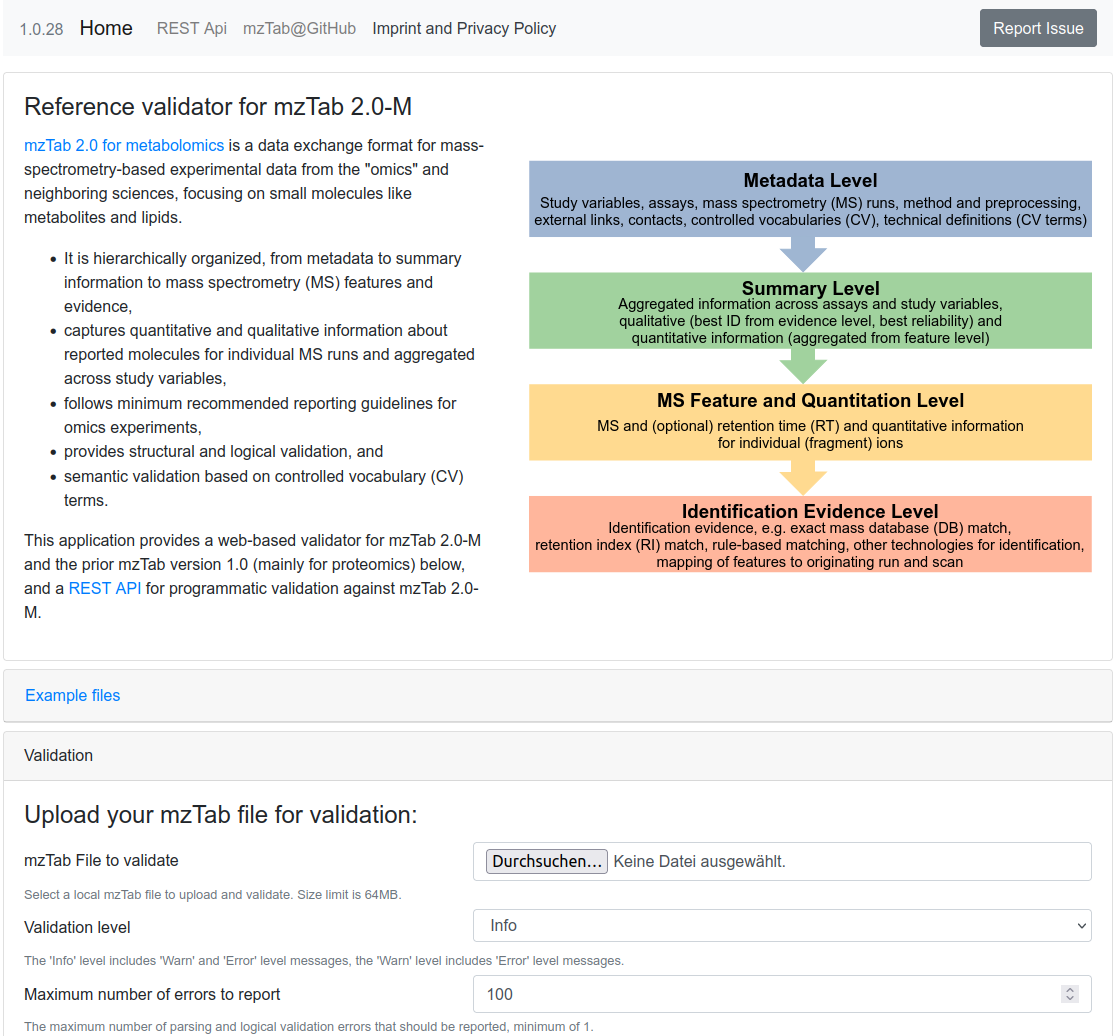
mzTab Validator
mzTab 2.0 for metabolomics is a data exchange format for mass-spectrometry-based experimental data from the "omics" and neighboring sciences, focusing on small molecules like metabolites and lipids. This application provides a web-based validator for mzTab 2.0-M and the prior mzTab version 1.0 (mainly for proteomics), and a REST API for programmatic validation against mzTab 2.0-M.


