Reference validator for mzTab 2.0-M
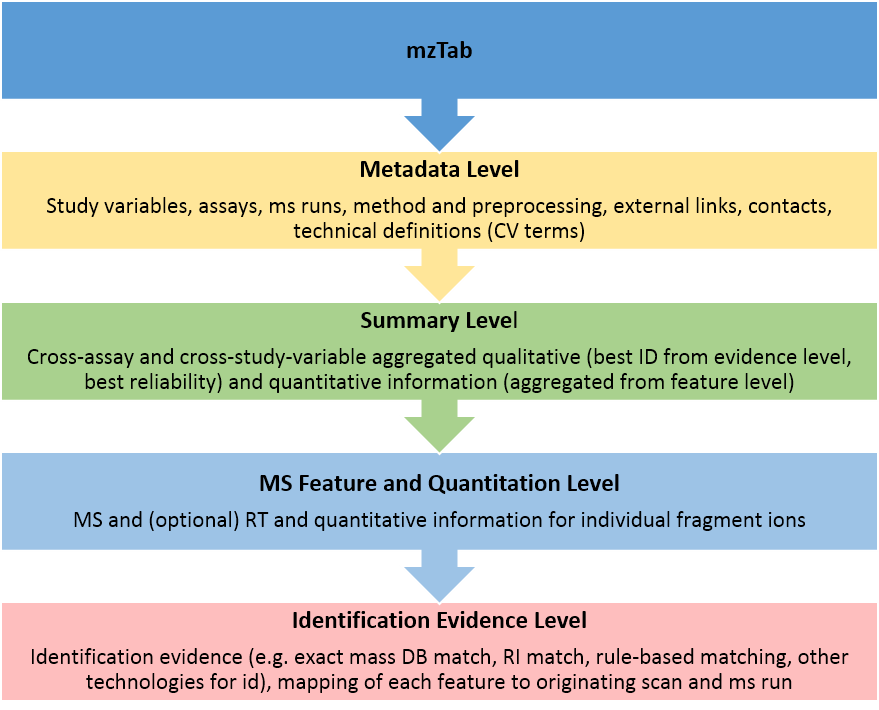
mzTab 2.0 for metabolomics is a data exchange format for mass-spectrometry-based experimental data from the "omics" and neighboring sciences, focusing on small molecules like metabolites and lipids.
- It is hierarchically organized, from metadata to summary information to ms features and evidence,
- captures quantitative and qualitative information about reported molecules for individual ms runs and aggregated across study variables,
- follows minimum recommended reporting guidelines for omics experiments, and
- provides structural, logical and semantic (controlled vocabulary terms) validation.
This application provides a web-based validator for mzTab 2.0-M and the prior mzTab version 1.0 (mainly for proteomics) below, and a REST API for programmatic validation against mzTab 2.0-M.
It is based on the jmzTab-M reference implementation.
