LipidXplorer msconvert conversion
MSConvert
Only a few mass spectrometers directly produce mzXML/mzML files but there are several tools available that generate mzXML/mzML files from native acquired files.
MSConvert is the currently most featured converter (MSConvert) and further it is under ongoing maintenance. It supports various vendor file formats:
- Agilent Mass Hunter Data Access Component Library
- Waters Raw Data Access Component Library
- Bruker CompassXtract
- Thermo-Scientific MSFileReader Library
- Waters Raw Data Access Component Library
- AB Sciex WIFF Reader Library
Download and installation
Download MSConvert from MSConvert. It is part of Protewizard, a tool for proteomics. Just click on the button "I agree to the license terms, download Proteowizard" and install the downloaded file.
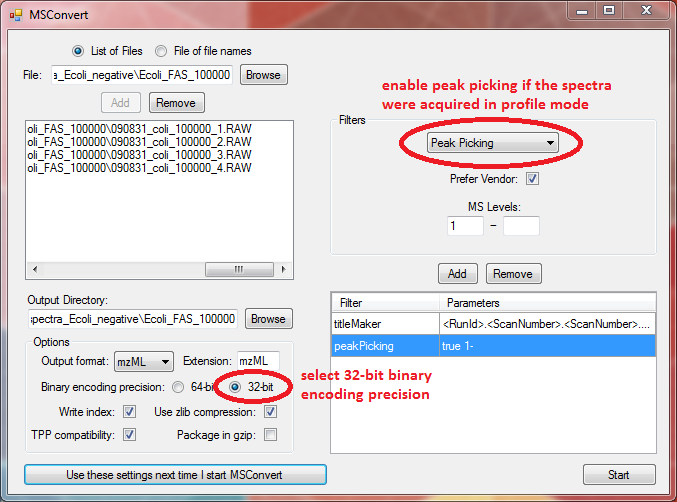
For LipidXplorer set the options in MSConvert like this:
For convenience store your settings by clicking "Use these settings next time I start MSConvert". Then upload your file(s) using "Browse" and "Add", respectively.
Use "Start" to do the conversion.
Always select 32-bit binary encoding precision.
Important: If you have your spectra imported in profile mode you have to convert it to a centroid spectrum by peak picking. To switch on the peak picking select it from the filters on the upper right. Select the MS level 1- and click "Add" (see screenshot above). Now the peak picker is enabled.