FAQ
LipidXplorer does not run although I have Python already on my computer
It can be that you already have Python installed but not 2.6 or 2.7. In this case, you have to install 2.6 or 2.7 (see LipidXplorer Installation). If you have one of those version make sure that you installed all the dependencies (see LipidXplorer Installation).
If you happen to have several versions of Python installed, there are different possibilities of how to solve this. On Windows the order of the Python exe in the PATH variable defines the version which is used when running LipidXplorer. Make sure that version 2.6 or 2.7 is the most left. But the best way is to explicitly call Python from the command line. On Windows this could be for example:
C:\Users\Herzog\LipidXplorer>c:\Python27\python.exe LipidXplorer.py
Here, LipidXplorer is located in c:\Users\Herzog.
On Linux one has to start LipidXplorer with the correct version which should be found somewhere like:
/usr/local/bin/python-2.6
Python is installed, LipidXplorer is installed, but clicking on lpdxGUI.pyw does nothing
Right click on LipidX or lpdxGUI.pyw. In the upcoming menu select Edit with IDLE. This should open 2 windows. One is the Python Shell and the other is the source code of lpdxGUI.pyw. Click on the source code window and press F5. If LipidX won't start then there will be an error message:
ImportError: DLL load failed: This application has failed to start because the application configuration is incorrect. Reinstalling the application may fix this problem.:
- This error occurs because the C runtime library is not being installed correctly on your machine. (see [1]).
- Please install the following if you have a x32 processor or a x64 processor
When I click on a MFQL query in the LipidXplorer Wiki it opens a html page instead of the script
If you click on a *.mfql script the file menu opens first. Here you left-click again on the *.mfql script to finally open it. Or click with the right mouse button on it to "Save [it] As..." .
LipidXplorer does not find any lipid although there are for sure lipids in the sample
If your import file format is *.dta/*.csv check:
- if you acquired your sample in negative ion mode you have to add the preposition 'neg_' on your data folder. (see Reference)
Why are +-0.5 in the double bond equivalents?
LipidXplorer calculates the double bond equivalent with a mathematical formula. If it is an ion, it has one proton less or more. This gives a 0.5 in the calculation. In more detail:
- +0.5 if the ion is negative
- -0.5 if the ion is positive
Is there a difference in quality between spectra recorded in 'profile' mode and 'centroid' mode?
LipidXplorer offers the conversion of profile mode recorded spectra into centroided spectra using readw.exe ([ReAdW]). First of all, we offered this feature to support a broader spectrum of input files. But we also discovered sometimes differences in quality from the spectra acquisition. On some low resolution mass spectrometers the quality of the 'centroid' recorded spectra is higher than the recording of 'profile' spectra with subsequent conversion into centroid.
What is the ‘dump file’ and how is it created?
Please have look into Reference, section 'The Run panel'
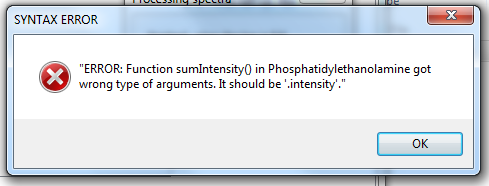
I imported all spectra, but when I want to identify lipids I get this error
The reason for this error is an old version of a query (which is in this case 'Phospahtidylethanolamine'). In the REPORT section is the function "sumIntenstiy()". It has arguments like this
FASINTENS = sumIntensity(FA1, FA2);
and should be changed to
FASINTENS = sumIntensity(FA1.intensity, FA2.intensity);