LipidXplorer Tutorial
Tutorial
Prerequisite
Install LipidXplorer as described in LipidXplorer_Installation. The LipidXplorer installer will create an icon on the desktop with which the programm can be started.
Example spectra are provided here:
Open the *.zip file and store the content of spectra_tutorial\spectra_Ecoli_negative\* to a place of your choice. LipidXplorer cames with a suggested folder structure. You can store the spectra under LipidXplorer\spectra\negative\
The LipidXplorer workflow
The workflow of LipidXplorer is quite simple:
- all spectra of all the acquisitions are imported #LipidXplorer Import and stored in the MasterScan
- MFQL queries are loaded into LipidXplorer and run #LipidXplorer Run panel on the MasterScan
- the result can be viewed #LipidXplorer ouptut
The LipidXplorer graphical user interface
LipidXplorer comes with a comfortable graphical user interface. It is divided into 4 parts:
- Import Source
- Import Settings
- Run
- MS tools
The Import Source, Import Settings and Run tab are exactly for the first and second steps of the workflow. The MS tools tab is an extra tab with which the user can do some calculations.
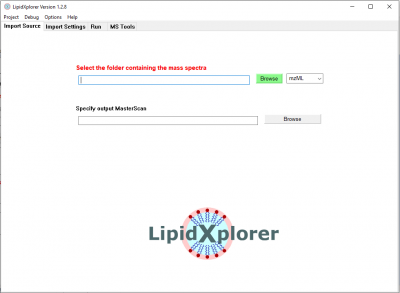
Specify input spectra
Open LipidXplorer and you will see the 'Import Source' panel. Click on the green 'Browse' button and select the folder
- LipidXplorer\spectra\negative\spectra_EColi_negative\spec_neg_100000 or
- <your chosen folder>\spectra_EColi_negative\spec_neg_100000 respectively
This folder contains 4 acquisitions of E.coli on an LTQ Orbitrap with high resolution MS and low resolution MS/MS spectra in negative mode.
Now the input folder with the spectra is selected. Make sure to have 'mzXML' selected next to the 'Browse' button, because the spectra are in mzXML format.
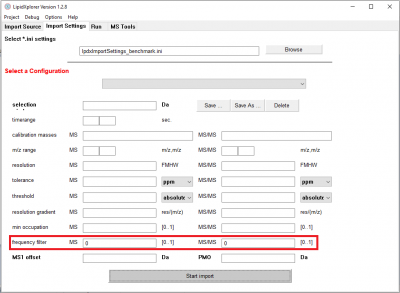
Specify the import settings
Go to the panel Import Settings.
For every import, machine-specific settings have to be set. LipidXplorer 1.2.8 offers new import settings which are the MSFilter and MSMSFilter. The MSFilter and MSMSFilter take a value between 0 and 1 that can be chosen to represent the fraction of scans in which a certain peak has to be present. The filter is applied for both MS and MS/MS according to the given import setting for each scan type. For each precursor ion, all associated MS/MS scans are collected according to the selection window. The counting of MS/MS scans and the association of the precursor are done automatically. Accordingly, the number of MS/MS scan can vary for each precursor ion in the shotgun acquisition, as might occur when using a DDA strategy. Moreover, the frequency-filter itself leads to a consistent intensity report for each acquisition file and in the aligned masterscan. In this perspective, one has to expect a reduction in a number of lipid identifications without losing abundant lipids.
All settings together can be stored in the file given at the top of the panel. 'lpdxImportSettings.ini' is the standard file in which a small collection of configuration are already stored. Select the configuration 'FAS_LTQ_100000MS1' under 'Select a Configuration'. These are the appropriate settings for the selected spectra.
Now hit 'Start Import' on the bottom of the panel and the import will start.
The debug window will report what LipidXplorer is doing.
When the import is ready, a small window will pop-up saying 'Task completed'.
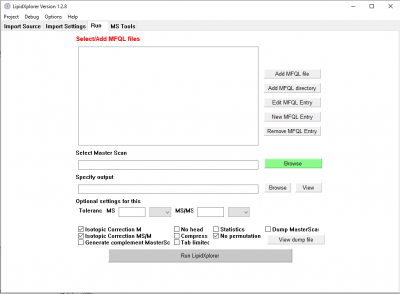
Identify lipids
Go to the panel Run.
Here we select the MFQL queries we want to use for the identification. Click on 'Add MFQL directory' to select all MFQL files of a folder. Select
- LipidXplorer/mfql/negative_mode.
This is a collection of queries for spectra in negative mode with MS/MS data. Once selected the queries are listed in the query window. You can also drag and drop folder or files there. A double click on of the queries will open a new panel with the content of the query. Here it can be edited.
Switch on the isotopic correction by clicking on 'Isotopic Correction MS' and 'Isotopic Correction MS/MS'. With a click on 'Run LipidXplorer' the software starts identification of lipids according to the loaded queries.
View the result
By clicking on 'View' (under the green 'Browse' button) the result will pop up as a table. It is automatically stored in the folder with the mass spectra. If you like to store it somewhere else under a different name click on 'Save as ...' in the result window.
The result can be viewed and edited with Excel.
Alternative import settings file
Please download the file File:LpdxImportSettings alternative.ini if you are going to work through the Basic Protocol 6 for Troubleshooting lipid identifications of
Herzog R, Schwudke D, Shevchenko A:LipidXplorer: Software for Quantitative Shotgun Lipidomics Compatible with Multiple Mass Spectrometry Platforms Current Protocols in Bioinformatics 2013 Oct 15